More Information
Submitted: July 22, 2025 | Approved: October 25, 2025 | Published: October 27, 2025
How to cite this article: Li J, Cai Y, Zhu S, Lin L, Zhang ZL, Xu QF, et al. Association of EIF4G1 Gene Variants with Sporadic Parkinson’s disease in a Chinese Han Population. J Neurosci Neurol Disord. 2025; 9(2): 067-071. Available from:
https://dx.doi.org/10.29328/journal.jnnd.1001112
DOI: 10.29328/journal.jnnd.1001112
Copyright License: © 2025 Li J, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Eukaryotic translation initiation factor 4G1(EIF4G1); Parkinson’s disease; rs2178403; mTOR signaling pathway; α-synuclein aggregation; Proteostasis dysregulation
Association of EIF4G1 Gene Variants with Sporadic Parkinson’s disease in a Chinese Han Population
Jin Li1*, Yu Cai2#, Si Zhu1#, Lu Lin3, Zhi-Ling Zhang1* and Qi-Feng Xu1*
1Department of Neurology, The First Affiliated Hospital of Guangzhou Medical University, Guangzhou 510120, China
2Guangzhou Key Laboratory of Spine Disease Prevention and Treatment, Department of Orthopaedic Surgery, The Third Affiliated Hospital of Guangzhou Medical University, Guangzhou, 510150, China
3Department of Neurology, the First Affiliated Hospital of Gannan Medical University, Ganzhou, 341000, China
#These authors contributed equally
*Address for Correspondence: Qi-Feng Xu, Department of Neurology, The First Affiliated Hospital of Guangzhou Medical University, Guangzhou 510120, China, Email: [email protected]
Zhi-Ling Zhang, Department of Neurology, The First Affiliated Hospital of Guangzhou Medical University, Guangzhou 510120, China, Email: [email protected]
Eukaryotic translation initiation factor 4G1 (EIF4G1) has been implicated in Parkinson’s disease (PD) pathogenesis. However, the contribution of EIF4G1 genetic variation to PD susceptibility remains unclear. To investigate the association between the EIF4G1 variant rs2178403 and PD risk. We analyzed EIF4G1 expression in PD and control samples using public GEO datasets (GSE54536). Additionally, we conducted a hospital-based case-control study with 541 sporadic PD patients and 401 age-/sex-matched healthy controls of Han Chinese ancestry. Genotyping of rs2178403 was performed using Sequenom MassARRAY iPLEX. GEO data revealed a non-significant trend toward elevated EIF4G1 expression in PD samples (p < 0.1). Genetic analysis identified a significant association between the rs2178403 GG genotype and increased PD risk under a recessive model (OR = 1.31, 95% CI = 1.010–1.703, p = 0.042). Stratified analysis showed a stronger effect in females. These findings suggest rs2178403 may contribute to PD susceptibility in the Han Chinese population. This study supports an association between the EIF4G1 variant rs2178403 and PD risk. Further investigation into EIF4G1 inhibition as a potential therapeutic strategy for PD is warranted.
Parkinson’s disease (PD) is the second most prevalent neurodegenerative disorder, affecting approximately 1% of individuals over 60 years and 4% - 5% of those over 85 [1,2]. PD is pathologically characterized by the progressive loss of dopaminergic neurons in the substantia nigra and the presence of Lewy bodies containing aggregated α-synuclein [3,4]. Clinically, PD presents with motor symptoms such as tremors, rigidity, bradykinesia, and postural instability, alongside non-motor symptoms like depression, sleep disturbances, and cognitive decline [5]. The etiology of PD is multifactorial, involving a complex interplay of genetic predispositions, environmental exposures, and aging-related cellular changes [6,7].
Recent genetic studies have identified both familial and sporadic forms of Parkinson’s disease (PD), with numerous risk loci implicated through genome-wide association studies (GWAS) [8]. Among molecular pathways linked to PD pathogenesis, the mammalian target of rapamycin (mTOR) signaling cascade has garnered significant attention due to its central role in regulating autophagy, protein synthesis, and cellular metabolism [9,10]. mTOR functions through two multiprotein complexes, mTORC1 and mTORC2, with well-characterized upstream regulators and downstream effectors [11].
A key mTORC1 substrate is the eukaryotic translation initiation factor 4E-binding protein family (EIF4E-BP1/2/3) [12]. When hypophosphorylated, EIF4E-BPs sequester eIF4E and prevent assembly of the eIF4F complex, thereby suppressing cap-dependent translation initiation. The scaffolding protein EIF4G1 is essential for eIF4F complex formation, which includes eIF4E and eIF4A [13]. The eukaryotic translation initiation factor 4G1 (EIF4G1) is a scaffolding protein essential for assembling the eIF4F complex, which includes eIF4E and eIF4A [14,15]. Under normal physiological conditions, EIF4G1 interacts with eIF4E to recruit ribosomes to the 5’-cap of mRNAs, thus initiating protein synthesis [16,17]. However, alterations in EIF4G1 levels or function can disturb proteostasis and have been associated with various neurodegenerative diseases [18]. Dysregulation of EIF4G1 expression or function disrupts proteostasis and is implicated in neurodegenerative disorders. In PD, impaired mTORC1-eIF4F signaling contributes to the accumulation of misfolded α-synuclein by compromising both its clearance and the translation of proteostatic machinery [19].
Although EIF4G1 mutations have been reported in familial PD, primarily in European cohorts, their role in sporadic PD across diverse ethnic populations remains underexplored [20]. To address this gap, we investigated the association between the EIF4G1 variant rs2178403 and PD risk in a Han Chinese population. Complementarily, we analyzed EIF4G1 expression patterns in public transcriptomic datasets to assess functional relevance. This integrated genetic-transcriptomic approach aims to clarify the contribution of EIF4G1 to PD susceptibility and provide a mechanistic foundation for future therapeutic targeting.
Gene expression analysis
Gene expression data from Parkinson’s disease (PD) patients and healthy controls were retrieved from the Gene Expression Omnibus (GEO) database (dataset GSE54536, designated as the training cohort). Following standardization of all samples, differential expression analysis was performed using thresholds of |log₂ fold change| > 1.5 and false discovery rate (FDR) < 0.1.
Study population
Our case–control study recruited a total of 942 Han Chinese subjects, including 541 sporadic PD patients and 401 healthy subjects matched by age, sex, and ethnicity. Participants were recruited from the Parkinson Clinic Center of the First Affiliated Hospital of Sun Yat‐sen University from January 2014 to June 2016. Parkinson’s disease was diagnosed in accordance with the UK PD Society Brain Bank clinical diagnostic criteria [21]. The study was approved by the Ethics Committee of the First Affiliated Hospital of Sun Yat‐Sen University, and all study subjects provided their informed written consent.
DNA isolation and single-nucleotide polymorphism genotyping
DNA extraction used the standard phenol-chloroform method [22]. Rs2178403 genotyping employed Sequenom MassARRAY iPLEX (San Diego, CA).
Statistical analysis
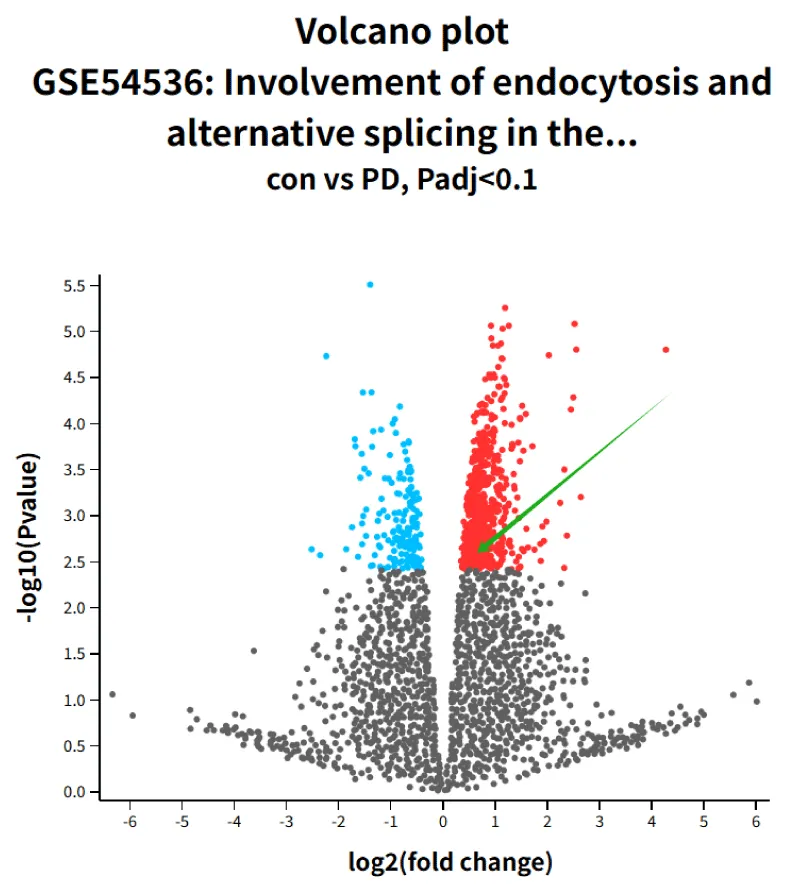
Our data comes from Gene Expression Omnibus (GEO) datasets (GSE54536). A volcano plot was used to identify differentially expressed genes (using n-fold ≥1.5 and p - value < 0.1 as the threshold of statistical significance).
The haplotype block was analyzed with Haploview and used Beagle 4.1 for further haplotype analysis. For all other statistical analyses, we used SPSS, version 27.0 (IBM Corp., Armonk, NY, USA) [23]. Genotypes and allele frequencies were determined by direct counting. Sex and age differences between patients and controls were examined using Student’s t‐test. The Hardy–Weinberg equilibrium (HWE) of the genotype distributions across patients and controls was calculated using a chi‐squared test. To test for differences in genotype and allele distributions between patients and controls, we used chi‐squared tests or Fisher’s exact tests. The odds ratio (OR) and the 95% confidence interval (CI) were used to describe the association between SNPs and the PD risk using logistic regression adjusted for age and sex. p < 0.05 (two‐tailed) was considered significant.
EIF4G1 showed a more significant increase in the PD group than control
We screened 874 differentially expressed genes (DEGs) between the PD group and the control group in GSE54536 using the R package “limma” (p < 0.1). The analysis data show that EIF4G1 has a higher increase in the PD group than control. The green arrow in group 1 points to the position of EIF4G1.
Characteristics of the study population
Our case–control study enrolled a total of 942 Han Chinese participants. The PD group included 541 subjects (320 men and 221 women). The control group included 401 age‐, sex‐, and ethnicity‐matched subjects (229 males and 172 females). The details are outlined in Table 1.
| Table 1: Demographic characteristics. | |||
| PD group (n = 541) |
Control group (n = 401) |
p | |
| Sex (male/female) | 320/221 | 229/172 | 0.530 |
| Age(mean±SD) | 63.36±10.74 | 62.88±10.13 | 0.938 |
Distributions of the rs2178403 polymorphism in PD and control groups
The genotype distribution of rs2178403 in both the PD and healthy control groups was determined to identify any deviation from HWE. Genotype and allele frequencies for rs2178403 are summarized in Table 2.
| Table 2: The genotype and allele frequencies for the SNP. | |||||||
| Group | Genotype | Allele | HWE p |
||||
| GG(%) | AG(%) | AA(%) | G(%) | A(%) | |||
| rs2178403 | PD(n = 541) | 256(47.32) | 235(43.44) | 50(9.24) | 747(69.04) | 335(30.96) | 0.708 |
| Control(n = 401) | 163(40.65) | 186(46.38) | 52(12.97) | 512(63.84) | 290(36.16) | 0.926 | |
| female | PD(n = 221) | 114(51.59) | 82(37.10) | 25(11.31) | 310(70.14) | 132(29.86) | |
| Control(n = 179) | 63(35.20) | 84(46.93) | 25(13.97) | 210(61.05) | 134(38.95) | ||
| male | PD(n = 320) | 142(44.38) | 153(47.81) | 25(7.81) | 437(68.28) | 156(24.37) | |
| Control(n = 229) | 100(43.67) | 102(44.54) | 27(11.79) | 302(65.94) | 156(34.06) | ||
rs2178403 polymorphism and PD susceptibility
For rs2178403, GG genotype frequency in the PD group was significantly higher than in the control group, when compared by logistic regression analysis using recessive model and allelic models(AA +AG versus GG; OR =1.31, 95% CI = 1.010–1.703, p = 0.042; G versus A, OR =1.263, 95% CI = 1.041–1.532, p = 0.018). By contrast, no significant difference was identified using either the dominant (AA versus AG+GG, OR =1.46, 95% CI = 0.969–2.209, p = 0.069) (Table 3).
| Table 3: Summary of comparisons stratified by sex and age. | |||||
| Models | PD | Control | OR (95% CI) | p | |
| rs2178403 | Dominant (AA/(AG+GG) | 50/491 | 52/349 | 1.46(0.969-2.209) | 0.069 |
| Recessive (AA+AG)/GG) | 285/256 | 238/163 | 1.31(1.010-1.703) | 0.042 | |
| Allele (G/A) | 747/335 | 512/290 | 1.263(1.041-1.532) | 0.018 | |
| female | Dominant (AA/(AG+GG) | 25/196 | 25/147 | 1.33(0.73-2.42) | 0.343 |
| Recessive (AA+AG)/GG) | 107/114 | 109/63 | 1.84(1.23-2.78) | 0.003 | |
| Allele (G/A) | 310/132 | 210/134 | 1.50(1.11-2.02) | 0.008 | |
| male | Dominant (AA/(AG+GG) | 25/295 | 27/202 | 1.58(0.89-2.81) | 0.119 |
| Recessive (AA+AG)/GG) | 178/142 | 129/100 | 1.03(0.73-1.45) | 0.869 | |
| Allele (G/A) | 437/156 | 302/156 | 1.45(1.11-1.89) | 0.005 | |
Next, we compared both groups by subgroup analysis based on sex. We found that female patients with the recessive model and allelic models were associated with an increased risk of developing PD compared to female controls (AA +AG versus GG; OR = 1.84, 95% CI = 1.23–2.78, p = 0.003; G versus A, OR = 1.50, 95% CI = 1.11–2.02, p = 0.008). In addition, male patients with the allelic models were associated with an increased risk of developing PD compared to male controls (G versus A, OR = 1.45, 95% CI = 1.11–1.89, p = 0.005). Furthermore, the dominant models detected no difference in the analysis (Figure 1). The results of different comparisons are presented in Table 3.
Figure 1: GROUP 1 volcano plot of EIF4G1.
EIF4G1 was confirmed as a candidate PD gene by Chartier-Harlin, et al. [24] This case-control study investigated the association between the EIF4G1 variant rs2178403 and PD susceptibility in a Han Chinese cohort (541 patients and 401 controls). We concurrently analyzed EIF4G1 expression patterns in public datasets to assess functional relevance. Our results support growing evidence implicating dysregulated mRNA translation initiation via the mTORC1-eIF4F axis in PD pathogenesis.
Progressive dopaminergic neuron loss and α-synuclein aggregation are core hallmarks of Parkinson’s disease (PD), which reflect underlying proteostasis failure [4]. Parkinson’s disease is considered to have multiple causes, resulting from both genetic and non-genetic factors. Genetic variants with large effect sizes have been identified in approximately 20% of persons with Parkinson’s disease (monogenic Parkinson’s disease). Autosomal dominant Parkinson’s disease with incomplete penetrance includes variants in LRRK2, GBA1, VPS35, and SNCA [25,26]. mTORC1 signaling coordinates key cellular processes, including metabolism, autophagy, and cap-dependent translation through effectors such as EIF4E-BPs and the eIF4F complex [27]. Within eIF4F, EIF4G1 acts as a critical scaffolding protein essential for complex assembly [28]. EIF4G1 expression is increased in different types of cancers [29,30]. Besides, increased EIF4G1 could promote the formation of tumor emboli by facilitating the translation of IRES-containing p120 mRNAs [30]. Research has shown that Parkinson’s Disease Genes VPS35 and EIF4G1 Interact Genetically and Converge on α-Synuclein [31]. Our study focused on EIF4G1, although prior research in Han Chinese populations has shown no association between the EIF4G1 variant rs2178403 and sporadic PD [32].
rs2178403 is an exonic variant in EIF4G1 in which methionine (ATG) is substituted with valine (GTG) [33]. Under a recessive model, genetic analysis identified a significant association between rs2178403 and increased PD risk (OR = 1.31, p = 0.042). Notably, this effect was significantly stronger in females (OR = 1.84, p = 0.003) than in males. This pronounced sexual dimorphism may reflect neuroprotective interactions between estrogen and mTOR signaling. Further transcriptomic analysis revealed altered EIF4G1 expression in PD peripheral blood samples compared to controls. These findings support the biological plausibility of EIF4G1 involvement in PD pathogenesis, where dysregulated expression might disrupt eIF4F stoichiometry and drive aberrant translation. Such disturbances could ultimately compromise proteostasis, potentially by impairing clearance of misfolded α-synuclein or promoting synthesis of aggregation-prone proteins, contributing to nigrostriatal vulnerability [34].
Several limitations should be noted. The study’s focus on a single variant (rs2178403) highlights the need for broader analysis across the EIF4G1 locus. Public expression datasets may be limited by tissue specificity and cohort heterogeneity. While the observed gender disparity is statistically robust, its mechanism requires validation in neuronal models that account for hormonal modulation of mTOR-EIF4G1 crosstalk. Future work must determine how the rs2178403 genotype or altered EIF4G1 expression affects eIF4F complex dynamics, translation fidelity, and α-synuclein metabolism.
In conclusion, this study identifies rs2178403 within EIF4G1 as a risk variant for Parkinson’s disease (PD) susceptibility in the Han Chinese population, revealing novel sexual dimorphism in its effect. The female-predominant risk pattern suggests potential modulation of PD pathogenesis through endocrine-mTOR pathway interactions, highlighting the need for sex-specific therapeutic exploration. Our findings reinforce dysregulated mTORC1-mediated translation as a contributor to proteostatic failure in PD. Future research should expand genetic analyses across EIF4G1 regulatory networks in diverse populations, establish functional consequences using neuron-specific models, and explore therapeutic modulation of this pathway to mitigate α-synuclein pathology.
Author contribution
QFX and ZLZ contributed to the study concept and design, and critical revision of the manuscript for intellectual content. JL, YC, and SZ contributed to the study concept and design, acquisition of the data, and analysis and interpretation of data. JL, YC, and LL recruited the subjects. SZ and LL obtained CSF samples. JL, YC, and ZLZ extracted DNA samples and performed the experiments. JL and YC analyzed the data. JL wrote the manuscript. The final manuscript was read and approved by all authors.
Data availability statement
The datasets analyzed during the current study are available from the corresponding authors upon reasonable request.
- Ye H, Robak LA, Yu M, Cykowski M, Shulman JM. Genetics and pathogenesis of parkinson’s syndrome. Annu Rev Pathol. 2023;18. Available from: https://doi.org/10.1146/annurev-pathmechdis-031521-034145
- Tysnes OB, Storstein A. Epidemiology of parkinson’s disease. J Neural Transm (Vienna). 2017;124(8). Available from: https://doi.org/10.1007/s00702-017-1686-y
- Haelterman NA, Yoon WH, Sandoval H, Jaiswal M, Shulman JM, Bellen HJ. A mitocentric view of parkinson’s disease. Annu Rev Neurosci. 2014;37. Available from: https://doi.org/10.1146/annurev-neuro-071013-014317
- Kalia LV, Lang AE. Parkinson's disease. Lancet. 2015;386(9996). Available from: https://doi.org/10.1016/s0140-6736(14)61393-3
- Borsche M, Pereira SL, Klein C, Grünewald A. Mitochondria and parkinson’s disease: clinical, molecular, and translational aspects. J Parkinsons Dis. 2021;11(1). Available from: https://doi.org/10.3233/jpd-201981
- Valente EM, Arena G, Torosantucci L, Gelmetti V. Molecular pathways in sporadic PD. Parkinsonism Relat Disord. 2012;18 Suppl 1. Available from: https://doi.org/10.1016/s1353-8020(11)70023-2
- Portela A, Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28(10). Available from: https://doi.org/10.1038/nbt.1685
- Kia DA, Zhang D, Guelfi S, Manzoni C, Hubbard L, Reynolds RH, et al. Identification of candidate parkinson disease genes by integrating genome-wide association study, expression, and epigenetic data sets. JAMA Neurol. 2021;78(4). Available from: https://doi.org/10.1001/jamaneurol.2020.5257
- Park JS, Choe K, Lee HJ, Park TJ, Kim MO. Neuroprotective effects of osmotin in parkinson’s disease-associated pathology via the AdipoR1/MAPK/AMPK/mTOR signaling pathways. J Biomed Sci. 2023;30(1). Available from: https://doi.org/10.1186/s12929-023-00961-z
- Lan AP, Chen J, Zhao Y, Chai Z, Hu Y. mTOR signaling in parkinson’s disease. Neuromolecular Med. 2017;19(1). Available from: https://doi.org/10.1007/s12017-016-8417-7
- Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–63. Available from: https://doi.org/10.1146/annurev.biochem.68.1.913
- Querfurth H, Lee HK. Mammalian/mechanistic target of rapamycin (mTOR) complexes in neurodegeneration. Mol Neurodegener. 2021;16(1):44. Available from: https://doi.org/10.1186/s13024-021-00428-5
- Le Bacquer O, Combe K, Patrac V, Ingram B, Combaret L, Dardevet D, et al. 4E-BP1 and 4E-BP2 double knockout mice are protected from aging-associated sarcopenia. J Cachexia Sarcopenia Muscle. 2019;10(3):696–709. Available from: https://doi.org/10.1002/jcsm.12412
- Li K, Tan G, Zhang X, Lu W, Ren J, Si Y, et al. EIF4G1 is a potential prognostic biomarker of breast cancer. Biomolecules. 2022;12(12). Available from: https://doi.org/10.3390/biom12121756
- Kim KQ, Nanjaraj Urs AN, Lasehinde V, Greenlaw AC, Hudson BH, Zaher HS. eIF4F complex dynamics are important for the activation of the integrated stress response. Mol Cell. 2024;84(11). Available from: https://doi.org/10.1016/j.molcel.2024.04.016
- Lama-Sherpa TD, Jeong MH, Jewell JL. Regulation of mTORC1 by the rag GTPases. Biochem Soc Trans. 2023;51(2). Available from: https://doi.org/10.1042/bst20210038
- Kim SH, Choi JH, Marsal-García L, Amiri M, Yanagiya A, Sonenberg N. The mRNA translation initiation factor EIF4G1 controls mitochondrial oxidative phosphorylation, axonal morphogenesis, and memory. Proc Natl Acad Sci U S A. 2023;120(25). Available from: https://doi.org/10.1073/pnas.2300008120
- Saini P, Rudakou U, Yu E, Ruskey JA, Asayesh F, Laurent SB, et al. Association study of DNAJC13, UCHL1, HTRA2, GIGYF2, and EIF4G1 with parkinson’s disease. Neurobiol Aging. 2021;100. Available from: https://doi.org/10.1016/j.neurobiolaging.2020.10.019
- Yu L, Hu X, Xu R, Zhao Y, Xiong L, Ai J, et al. Piperine promotes PI3K/AKT/mTOR-mediated gut-brain autophagy to degrade α-synuclein in parkinson’s disease rats. J Ethnopharmacol. 2024;322. Available from: https://doi.org/10.1016/j.jep.2023.117628
- Lesage S, Condroyer C, Klebe S, Lohmann E, Durif F, Damier P, et al. EIF4G1 in familial parkinson’s disease: pathogenic mutations or rare benign variants? Neurobiol Aging. 2012;33(9). Available from: https://doi.org/10.1016/j.neurobiolaging.2012.05.006
- Hughes AJ, Daniel SE, Kilford L, Lees AJ. Accuracy of clinical diagnosis of idiopathic parkinson’s disease: a clinico-pathological study of 100 cases. J Neurol Neurosurg Psychiatry. 1992;55(3). Available from: https://doi.org/10.1136/jnnp.55.3.181
- Wang P, Liu X, Ye Z, Gong B, Yang Y, Zhang D, et al. Association of IGF1 gene polymorphism with parkinson’s disease in a Han Chinese population. J Gene Med. 2017;19(4). Available from: https://doi.org/10.3109/13816810.2016.1145699
- Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81(5). Available from: https://doi.org/10.1086/521987
- Chartier-Harlin MC, Dachsel JC, Vilariño-Güell C, Lincoln SJ, Leprêtre F, et al. Translation initiator EIF4G1 mutations in familial parkinson disease. Am J Hum Genet. 2011;89(3):398–406. Available from: https://doi.org/10.1016/j.ajhg.2011.08.009
- Blauwendraat C, Nalls MA, Singleton AB. The genetic architecture of parkinson’s disease. Lancet Neurol. 2020;19(2):170–8. Available from: https://doi.org/10.1016/s1474-4422(19)30287-x
- Tanner CM, Ostrem JL. Parkinson’s disease. N Engl J Med. 2024;391(5):442–52. Available from: https://doi.org/10.1056/nejmra2401857
- Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168(6):960–76. Available from: https://doi.org/10.1016/j.cell.2017.02.004
- Deng H, Wu Y, Jankovic J. The EIF4G1 gene and parkinson’s disease. Acta Neurol Scand. 2015;132(2). Available from: https://doi.org/10.1111/ane.12397
- Tu L, Liu Z, He X, He Y, Yang H, Jiang Q, et al. Over-expression of eukaryotic translation initiation factor 4 gamma 1 correlates with tumor progression and poor prognosis in nasopharyngeal carcinoma. Mol Cancer. 2010;9:78. Available from: https://doi.org/10.1186/1476-4598-9-78
- Silvera D, Arju R, Darvishian F, Levine PH, Zolfaghari L, Goldberg J, et al. Essential role for eIF4GI overexpression in the pathogenesis of inflammatory breast cancer. Nat Cell Biol. 2009;11(7):903–8. Available from: https://doi.org/10.1038/ncb1900
- Dhungel N, Eleuteri S, Li LB, Kramer NJ, Chartron JW, Spencer B, et al. Parkinson’s disease genes VPS35 and EIF4G1 interact genetically and converge on α-synuclein. Neuron. 2023;111(1):138. Available from: https://doi.org/10.1016/j.neuron.2022.12.020
- Li K, Tang B, Guo J, Lou M, Lv Z, Liu Z, et al. Analysis of EIF4G1 in ethnic Chinese. BMC Neurol. 2013;13:38. Available from: https://doi.org/10.1186/1471-2377-13-38
- Zhao Y, Ho P, Prakash KM, Foo JN, Liu JJ, Au WL, et al. Analysis of EIF4G1 in parkinson’s disease among Asians. Neurobiol Aging. 2013;34(4):1311.e5–6. Available from: https://doi.org/10.1016/j.neurobiolaging.2012.09.003
- Khurana V, Peng J, Chung CY, Auluck PK, Fanning S, Tardiff DF, et al. Genome-scale networks link neurodegenerative disease genes to α-synuclein through specific molecular pathways. Cell Syst. 2017;4(2):157–70.e14. Available from: https://doi.org/10.1016/j.cels.2016.12.011